Getting Started¶
Download and Installation¶
This guide assumes that you have access to an administrator or sudoer account on a macOS or Linux system.
Getting Organized¶
Creating Enclosing Directory¶
Create a vivarium_work folder anywhere you like. But for installing
some third-party software, everything we do will occur inside this
folder.
Setting PYTHONPATH¶
Vivarium needs the root of the repository to be in your PYTHONPATH
environment variable so that Python can find Vivarium. To make this easy
to set, we suggest adding this line to your shell startup file:
alias pycd='export PYTHONPATH="$PWD:$PYTHONPATH"'
Now when you are about to work on Vivarium, navigate to the root of the
Vivarium repository (vivarium_work/vivarium) and run pycd in
your terminal. You will need to do this for each terminal window you
use.
Installing Dependencies¶
Below, we list the dependencies Vivarium requires, how to check whether you have them, how to install them, and in some cases, how to set them up for Vivarium. Make sure you have each of them installed.
Python 3¶
We wrote Vivarium in Python 3. While we did also write it to be Python-2-compatible, the Python Software Foundation has deprecated Python 2, so we strongly recommend using Python 3.
Check Installation
$ python --version
Python <version>
Make sure you see a version at least 3.6.
Install
Download the latest installer from the Python download page
Open JDK 8¶
Zookeeper and Kafka, which we will address later, require that you have Java installed.
Check Installation
$ java -version
java version <version> <date>
Ensure the version is at least 1.8.
Install
Download the latest JDK installer from the Java SE Downloads site. As of
writing the latest is Java SE 14. Download and run the appropriate
installer for your platform. Then you need to set the JAVA_HOME
environment variable, for instance by adding export
JAVA_HOME=$(/usr/libexec/java_home) to your startup shell file (e.g.
~/.bash_profile or ~/.profile).
Zookeeper and Kafka¶
Kafka is a message passing system that allows decoupling of message senders and message receivers. It does this by providing two abstractions, a Consumer and a Producer. A Consumer can subscribe to any number of “topics” it will receive messages on, and a Producer can send to any topics it wishes. Topics are communication “channels” between processes that otherwise do not need to know who is sending and receiving these messages. Vivarium uses Kafka to pass messages between actors, for example between a cell and its environment.
Kafka relies on Zookeeper, which synchronizes access to a hierarchy of key-value pairs called nodes. We like Kafka because it will let us distribute the model across computers in a server cluster, but you can also run a local Kafka instance for development.
Check Installation
For this guide, we will not install Kafka globally on your system.
Instead, we will store the Kafka program in vivarium_work and run
the executable directly. This means you almost certainly need to install
it, even if you use Kafka already.
Install
Download Kafka from the Apache Kafka site, choosing the latest version. This will give you a
.tgzarchive file that includes both Kafka and Zookeeper.Unarchive this file into
vivarium_workto create a folder likevivarium_work/kafka_2.11-2.0.0/. Your folder name will likely change slightly to match your version of Kafka.Create a shell script
vivarium_work/zookeeper.shwith the following content:#!/bin/bash ./kafka_2.11-2.0.0/bin/zookeeper-server-start.sh \ ./kafka_2.11-2.0.0/config/zookeeper.properties
Create a shell script
vivarium_work/kafka.shwith the following content:#!/bin/bash ./kafka_2.11-2.0.0/bin/kafka-server-start.sh \ ./kafka_2.11-2.0.0/config/server.properties \ --override listeners=PLAINTEXT://127.0.0.1:9092
Overriding the “listeners” address like this allows connections to the Kafka server to withstand network DHCP address changes and the like.
Make the scripts executable like this:
$ chmod 700 vivarium_work/kafka.sh $ chmod 700 vivarium_work/zookeeper.sh
Now you can start and stop the Zookeeper and Kafka servers like this:
$ vivarium_work/zookeeper.sh $ vivarium_work/kafka.sh
Make sure to start Zookeeper before Kafka, as Kafka expects a Zookeeper instance to already be running when in starts. Also note that you must run these two commands in separate terminals. To shut them down, you can just use CTRL-C to kill the processes.
Warning
Make sure you shut down Kafka before Zookeeper! If you shut down Zookeeper first, Kafka will refuse to quit. You can then force it to stop with
kill -9.
MongoDB¶
We use a MongoDB database to store the data collected from running simulations. This can be a remote server, but for this guide we will run a MongoDB server locally.
Check Installation
$ mongod --version
db version v4.2.3
...
Make sure you see a version at least 4.2.
Install
If you are on macOS, you can install MongoDB using Homebrew. You will need to add the MongoDB tap following the instructions here.
If you are on Linux, see the MongoDB documentation’s instructions.
Setup
You can get a MongoDB server up and running locally any number of ways. Here is one:
Create a folder
vivarium_work/mongodb. This is where MongoDB will store the database We store the database here instead of at the default location in/usr/local/var/mongodbto avoid permissions issues if you are not running as an administrator.Make a copy of the
mongodconfiguration file so we can make changes:$ cp /usr/local/etc/mongod.conf vivarium_work/mongod.confNote that your configuration file may be somewhere slightly different. Check the MongoDB documentation for your system.
In
vivarium_work/mongod.confchange the path afterdbPath:to point tovivarium_work/mongodb.Create a shell script
vivarium_work/mongo.shwith the following content:#!/bin/bash mongod --config mongodb.confMake the script executable:
$ chmod 700 vivarium_work/mongo.sh
Now you can launch MongoDB by running this script:
$ vivarium_work/mongo.sh
GNU Linear Programming Kit (GLPK)¶
One of the Python packages we will install later, swiglpk, requires
that you already have GLPK installed on your system.
Check Installation
We don’t have a way to check whether you have glpk installed. If you
think you already have it, you can proceed with the installation and
watch for an error about missing glpk.
Install
If you use Homebrew, you can install GLPK like this:
$ brew install glpk
Otherwise, follow the installation instructions on the GLPK homepage.
Leiningen¶
Our simulation runs each cell on its own thread, and we use Leiningen to manage these threads.
Check Installation
To check whether you have Leiningen installed, run:
$ lein --version
Leiningen <version> ...
You may also see a deprecation warning from Java HotSpot, which you can ignore. Make sure the version is at least 2.9.
Install
To install Leiningen, follow the instructions on its website. You can also install the leiningen
formula on Homebrew instead.
Download and Setup Vivarium¶
Download the Code¶
The Vivarium code is available on GitHub. Move into your
vivarium_work directory and clone the repository to
download the code
$ cd vivarium_work
$ git clone https://github.com/CovertLab/vivarium.git
This will create a vivarium folder inside vivarium_work. All the
code for Vivarium is inside this vivarium folder.
Installing Python Packages¶
Above we installed all the non-Python dependencies, but we still have to install the Python packages Vivarium uses.
Move into the
vivariumfolder created when you cloned the repository.(optional) Create and activate a virtual environment:
$ python3 -m venv venv ... $ source venv/bin/activate
Install Numpy. One of our dependencies,
stochastic-arrowrequires that Numpy be installed first. Check therequirements.txtfile for a line like this:numpy==1.15.3
Now install the version of Numpy specified in
requirements.txt$ pip install numpy==1.15.3
Install packages
$ pip install -r requirements.txtIf you encounter problems installing numpy and/or scipy, try this instead:
$ pip install -r requirements.txt --no-binary numpy,scipy $ pip install numpy $ pip install scipy
Now you are all set to run Vivarium!
Run Simulations¶
Some Terminology: Processes and Composites¶
In Vivarium, we break our cell models into processes. Each process models part of the cell’s function. For example, we have processes for metabolism, transcription, and translation in Vivarium. We can combine these processes into compartments that model a cell with all the functionality modeled by the included processes. For example, we could compose transcription and translation to create a fuller gene expression model.
In Vivarium, we store individual processes in
vivarium/vivarium/processes and compartments of processes in
vivarium/vivarium/compartments.
Running Processes and Composites in Isolation¶
You can run any process or compartment by itself. While this is too simple for modeling whole cells or colonies, simulating processes in isolation lets you study their dynamics and tune them to different conditions. To run a process or compartment, you can execute the Python file that defines it. For example, we can run the degradation process like this:
$ python vivarium_work/vivarium/vivarium/processes/degradation.py
...
Tip
If you get errors from Python about being unable to find
vivarium, make sure you’ve added the repository root to your
PYTHONPATH. See Setting PYTHONPATH for details.
Don’t worry about the output–it’s for developers. You will see a new
folder at vivarium_work/vivarium/vivarium/out/tests. This is where
we store the output from running processes and compartments in isolation.
For the degradation process, the output is in the degradation folder
inside tests. Here you’ll find a simulation.png file that looks
like this:
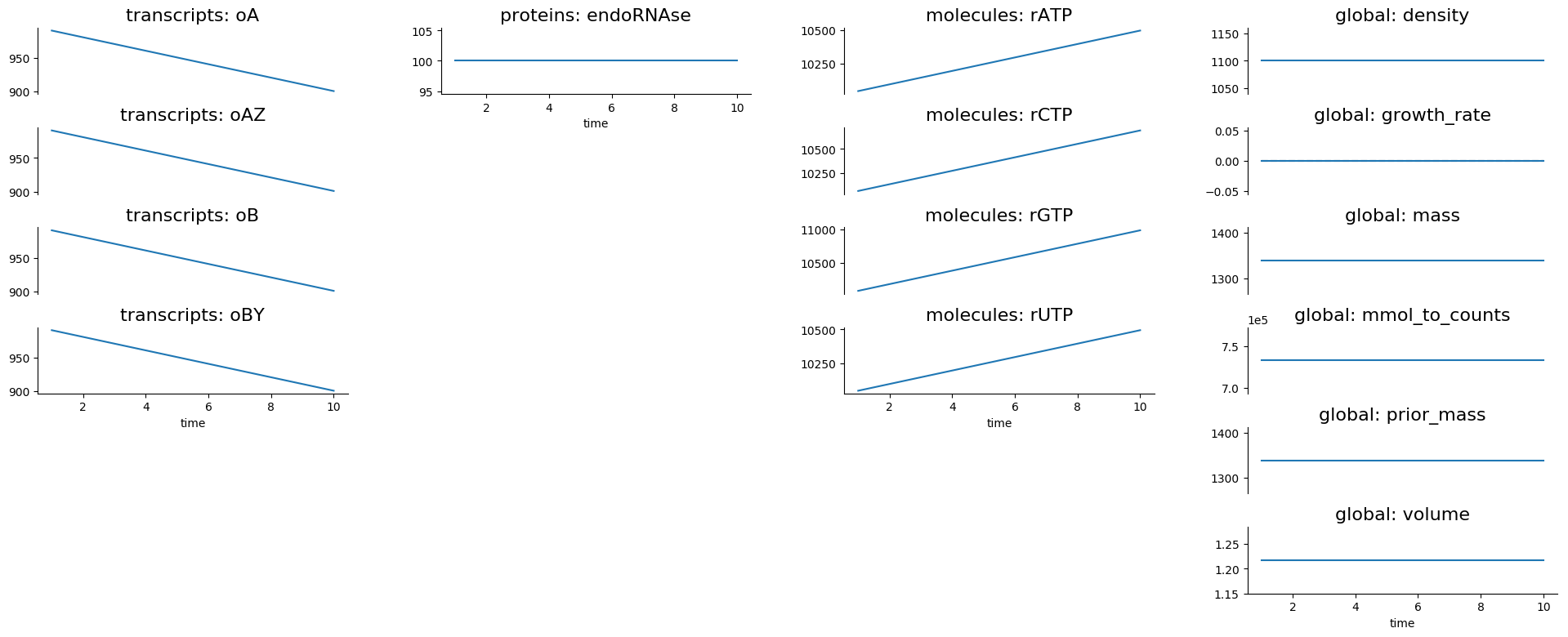
If you wanted to understand how the degradation process works, this would show you that it removes transcripts and returns the RNA nucleotides to the cell.
Some processes also produce the data shown in the plots. You can find
this data in simulation_data.csv. Try running the
convenience_kinetics process to see how this works!
Lastly, try running the flagella_expression compartment like this:
$ python vivarium_work/vivarium/vivarium/compartments/flagella_expression.py
Now in the flagella_expression_composite (the suffix _composite
is left over from when we called compartments “composites”) in
tests, you should see an image containing a plot like this:

Notice that even from this minimal simulation, we can tell which amino acid is limiting! In this case the colors are so similar that it’s hard to tell, but the limiting amino acid is either alanine or leucine.
Running Agents in Terminal Windows¶
Tip
Running agents separately in terminal windows is helpful for debugging because it lets you see the output from each agent.
Terminology: Agents¶
Vivarium is heavily influenced by agent-based modeling, in which the model consists of individual agents interacting with each other. In Vivarium, cells are agents that move around and grow within a shared environment. These agents interact with each other and their environment by passing messages through Kafka.
How to Run Agents¶
Each agent runs on its own thread. We do this because each agent can be as complex as an entire whole-cell model, so the entire simulation cannot run on a single thread. Shepherd can manage these threads for you; importantly, you must use Shepherd if your simulation will require creating or deleting threads. Cell division, for example, involves stopping the mother cell’s thread and starting two new threads, one for each daughter cell, so division requires Shepherd.
That said, you can run agents on your own instead of using Shepherd.
Warning
If you run a simulation using this method that includes stopping and/or starting agents, the agents will stop, but new ones will not start. For example if your cell divides, the agent you started for the mother cell will stop, but the daughter cells will not start.
We will run each agent in its own terminal window to mimic the threads that Shepherd would create. Let’s see how!
First we need to get all our servers running. Do each of the following in a separate terminal window:
Start Zookeeper:
$ vivarium_work/zookeeper.sh ... ... INFO binding to port 0.0.0.0/0.0.0.0:2181 ...
Start Kafka:
$ vivarium_work/kafka.sh ... ... INFO [KafkaServer id=0] started (kafka.server.KafkaServer)
You should also see som text print out on the Zookeeper window. You might see some
NoNodewarnings–these are safe to ignore.Warning
You must start Zookeeper before Kafka!
Start MongoDB:
$ vivarium_work/mongo.shThere shouldn’t be any output.
If you installed MongoDB using Homebrew, you can instead tell Homebrew to always run a MongoDB server by running:
$ brew services start mongodb/brew/mongodb-communityNow a MongoDB server will start automatically once you login. Then you can skip the step of starting MongoDB in the future.
Now we can create our agents. We create an agent like this:
$ python -m vivarium.environment.boot --type <type> --id <id> [--outer-id <outId>]
Tip
If you get errors from Python about being unable to find
vivarium, make sure you’ve added the repository root to your
PYTHONPATH. See Setting PYTHONPATH for details.
where <type> is the agent type, <id> is the identifier for this
agent, and <outId> is an optional argument that instructs Vivarium
to place the new agent inside the agent with identifier <outId>.
This outer agent will almost always be an environment. You can also
provide an optional --config '{...}' argument you can use to
configure the agent.
To see the other agent types, check out the help text like this:
$ python -m vivarium.environment.boot --help
Here’s an example of running a simulation of a simple environment with three cells that consume glucose and lactose. We will initialize the environment with glucose and lactose, and as the cells deplete the glucose we should see the cells shift to consuming lactose.
First, let’s create a
ecoli_core_glcenvironment agent. This is a kind of lattice environment. Lattice environments discretize the simulation space into a two-dimensional grid, each region of which has the same depth. Each region has uniform metabolite concentrations, but metabolite concentrations differ between regions, letting us model a continuous distribution of concentrations. A diffusion process in the environment tends to make the space homogeneous. We start this agent like this:$ python -m vivarium.environment.boot --type ecoli_core_glc --id env environment started
Warning
Wait for the
environment startedto show up before proceeding. Otherwise there won’t be an environment to add the cells to!Next, let’s create three cell agents. These agents will be of type
shifterbecause they will initially consume glucose, but when glucose concentrations drop, they will start consuming lactose. We create these agents like this:$ python -m vivarium.environment.boot --type shifter --id c1 --outer-id env $ python -m vivarium.environment.boot --type shifter --id c2 --outer-id env $ python -m vivarium.environment.boot --type shifter --id c3 --outer-id env
After creating each cell agent, you should see in both the cell and the environment’s terminal windows a message from the cell to the environment declaring itself:
<-- environment-receive CELL_DECLARE [shifter c1]: {'event': 'CELL_DECLARE', 'agent_id': 'env', 'inner_id': 'c1', 'agent_config': { ... }, 'state': {'volume': 1.0}}
And a message from the environment back to the cell:
<-- cell-receive ENVIRONMENT_SYNCHRONIZE [glc_lct env]: {'event': 'ENVIRONMENT_SYNCHRONIZE', 'inner_id': 'c1', 'outer_id': 'env', 'state': { ... }}
Now we can start the simulation!
$ python -m vivarium.environment.control run --id envSimulations stop on their own once the environment agent hits the end of its programmed timeline. You can also pause, run, and shutdown the simulation like this:
$ python -m vivarium.environment.control pause --id env $ python -m vivarium.environment.control run --id env $ python -m vivarium.environment.control shutdown
In this example, one of the cells tries to divide, halting the simulation. We’ll see later how to simulate division.
Once the simulation completes, run the analysis script to plot the data:
$ python vivarium/analysis/run_analysis -e envWhen the script completes, look for a folder named
envinvivarium_work/vivarium/outwith plots from your simulation.
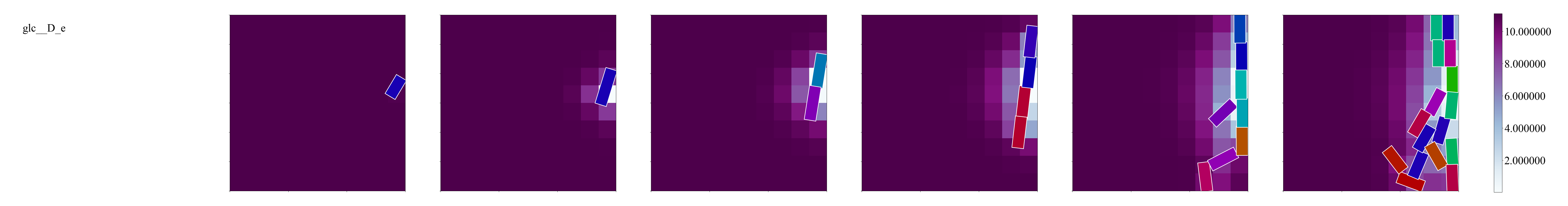
In snap_out.png you should see something like this:
Notice that the cells are consuming glucose and lactose as we expected!
Now take a look at c1/compartment.png. Here is part of the plot you
should see:
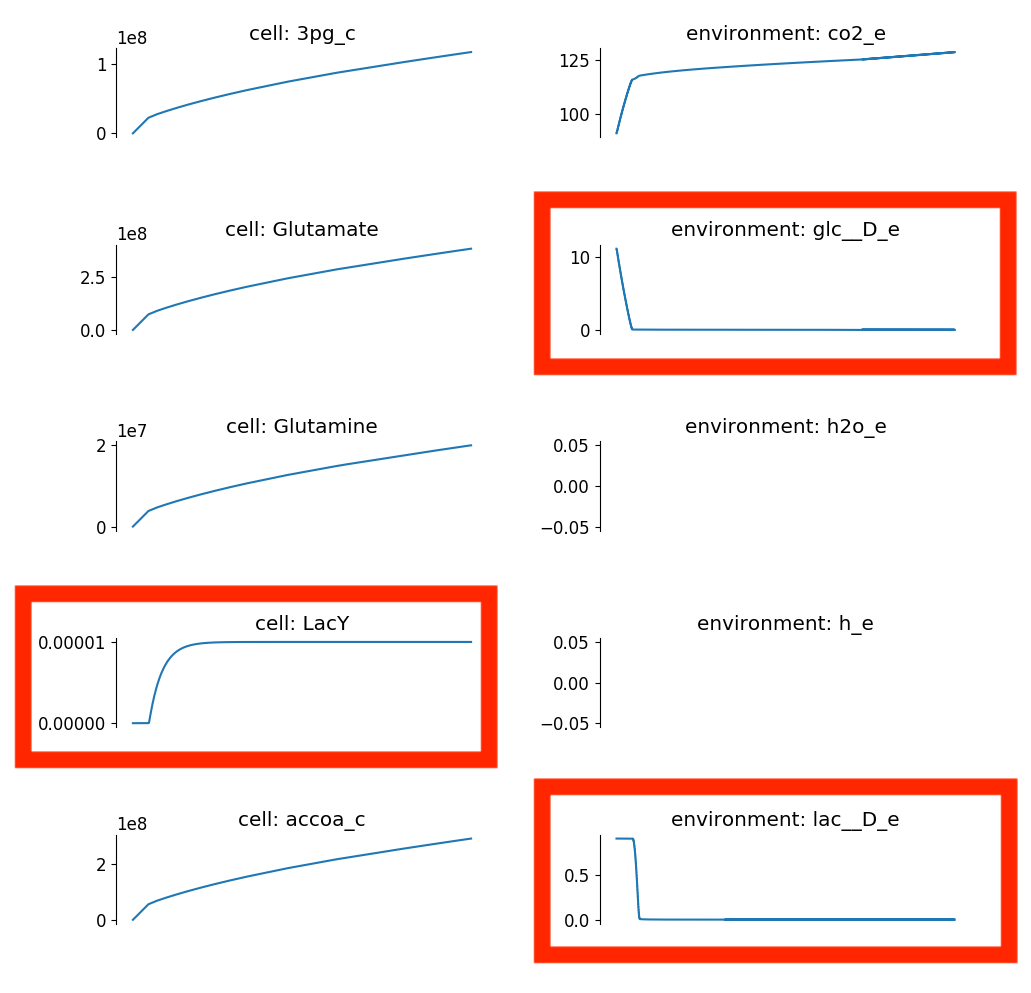
Here notice that the glucose levels near the cells drop precipitously, after which LacY expression increases. Then, the cell consumes the surrounding glucose, as we expected.
Using Shepherd¶
The usual way to start the simulation is to use Shepherd, which spawns agents in new threads as requested via Kafka messages so you don’t have to launch each agent in its own terminal tab. Furthermore, this enables cell division wherein a cell agent process ends and two new ones begin. To debug an agent, though, see the Running Agents in Terminal Windows instructions above.
Let’s take a look at an example of using Shepherd. We’ll be able to model cells dividing!
First, start Zookeeper, Kafka, and MongoDB as we discussed above in How to Run Agents.
Launch Shepherd in a separate terminal window:
$ lein runFor our environment, let’s make a
latticeagent:$ python -m vivarium.environment.boot --type ecoli_core_glc --id env2 environment started
Warning
Wait for the
environment startedto show up before proceeding. Otherwise there won’t be an environment to add the cells to!Tip
If you get errors from Python about being unable to find
vivarium, make sure you’ve added the repository root to your PYTHONPATH. See Setting PYTHONPATH for details.Next, let’s create a cell agent of type
growth_division, which can grow and divide.$ python -m vivarium.environment.boot --type growth_division --id c --outer-id env2Now we can start the simulation!
$ python -m vivarium.environment.control run --id env2This simulation is quite long, so feel free to cancel it with CTRL-C after you’re tired of waiting.
Once the simulation finishes, we can analyze the data:
$ python vivarium/analysis/run_analysis -e env2Note
You can run the analysis script while the simulation is still running too.
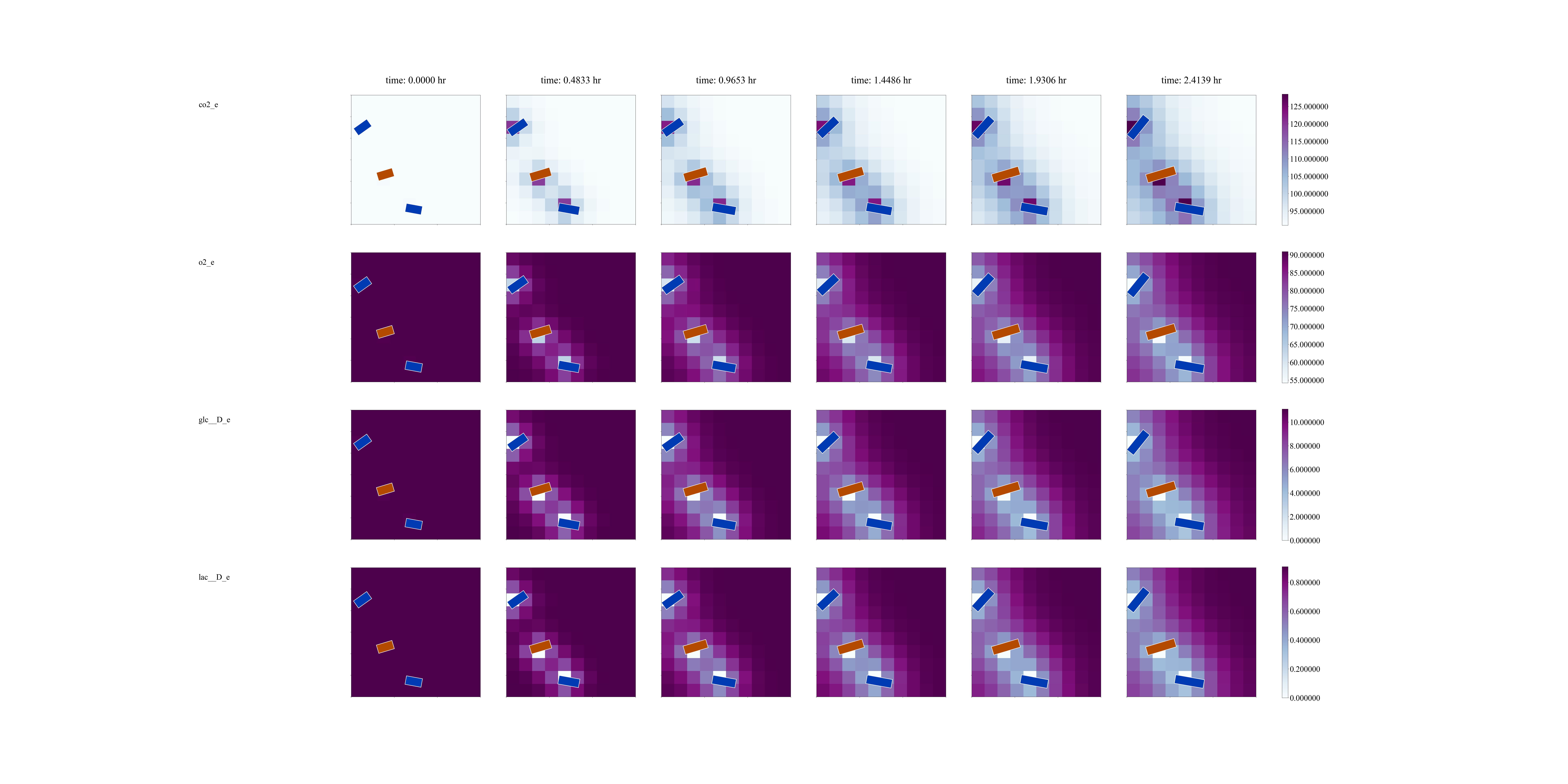
First, notice the folders in the analysis output. Each folder is a cell,
so since the cells divided, we have a lot of them! If you take a look at
snap_out.png, you should see rows of plots like this:

Running Experiments¶
With Shepherd, you can also run experiments that pre-define the environment and cell types. For example, let’s see how we could have run a simulation of growing and dividing cells like above with less work:
First, start Zookeeper, Kafka, and MongoDB as we discussed above in How to Run Agents.
Now start up Shepherd:
$ lein runLoad the experiment:
Danger
This experiment doesn’t work yet. We are working on a fix in #178
$ python -m vivarium.environment.control \ growth-division-experiment --experiment_id exp
Tip
If you get errors from Python about being unable to find
vivarium, make sure you’ve set your PYTHONPATH to include vivarium. See Setting PYTHONPATH for details.Run the simulation:
$ python -m vivarium.environment.control run --id expWhen it finishes, run the analysis:
$ python vivarium/analysis/run_analysis -e expThis is a long experiment, so you might want to end the simulation before it finishes.
In snap_out.png, we see a similar outcome to before. The plots are
different this time because there is some stochasticity in the model.