The Vivarium Modeling Paradigm¶
Our goal in building Vivarium is to help you leverage the molecular detail of whole-cell models in simulating the dynamics of populations of cells. To achieve this, we combined ideas from agent-based modeling, whole-cell modeling, and multiscale modeling.
An agent-based model contains simple agents whose interactions give rise to complex population dynamics. The behavior of the population is an emergent property, not one that is explicitly modeled. In Vivarium, we took inspiration from agent-based modeling in two major ways. First, we model cell populations by modeling individual cells and letting those cells interact in a shared environment. Second, we avoid specifying population behavior. Instead, we want realistic population dynamics to emerge from our modeling of the individual cells.
Unlike in agent-based modeling, Vivarium’s models of individual cells can have all the complexity of a whole-cell model. Whole-cell modeling simulates phenotypes at the scale of the cell by modeling the cell’s molecular mechanisms. Whole-cell models are also a hybrid modeling framework that integrates different mathematical representations of mechanism. In Vivarium, we model each of these molecular mechanisms by a process. We can then wire processes together to form compartments.
Since we are modeling phenomena that occur at spatial and temporal scales all the way from chemical reactions to cell population dynamics, we need to run our simulations at different scales as well. To do so, we draw from multiscale models, which contain sub-models that operate at different spatial and temporal scales. Some of these scales are illustrated below:
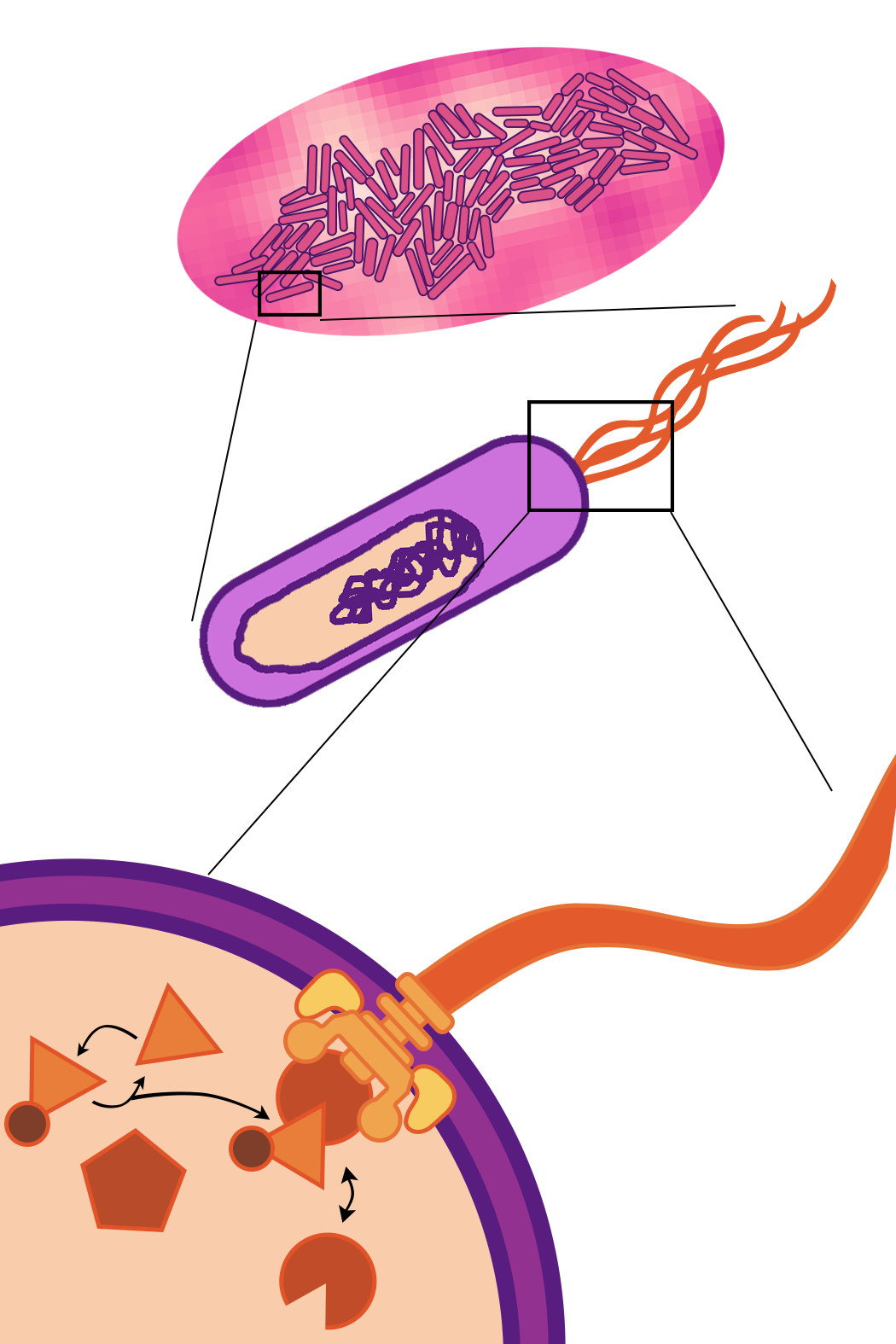
We describe temporal scales by timesteps, which define how finely we discretize time. Each process and each connection between them have a timestep.