Hierarchy¶
In Vivarium, we combine processes and stores to form compartments as shown in panel B below. These compartments are in turn nested to form the model hierarchy, depicted in panel C.
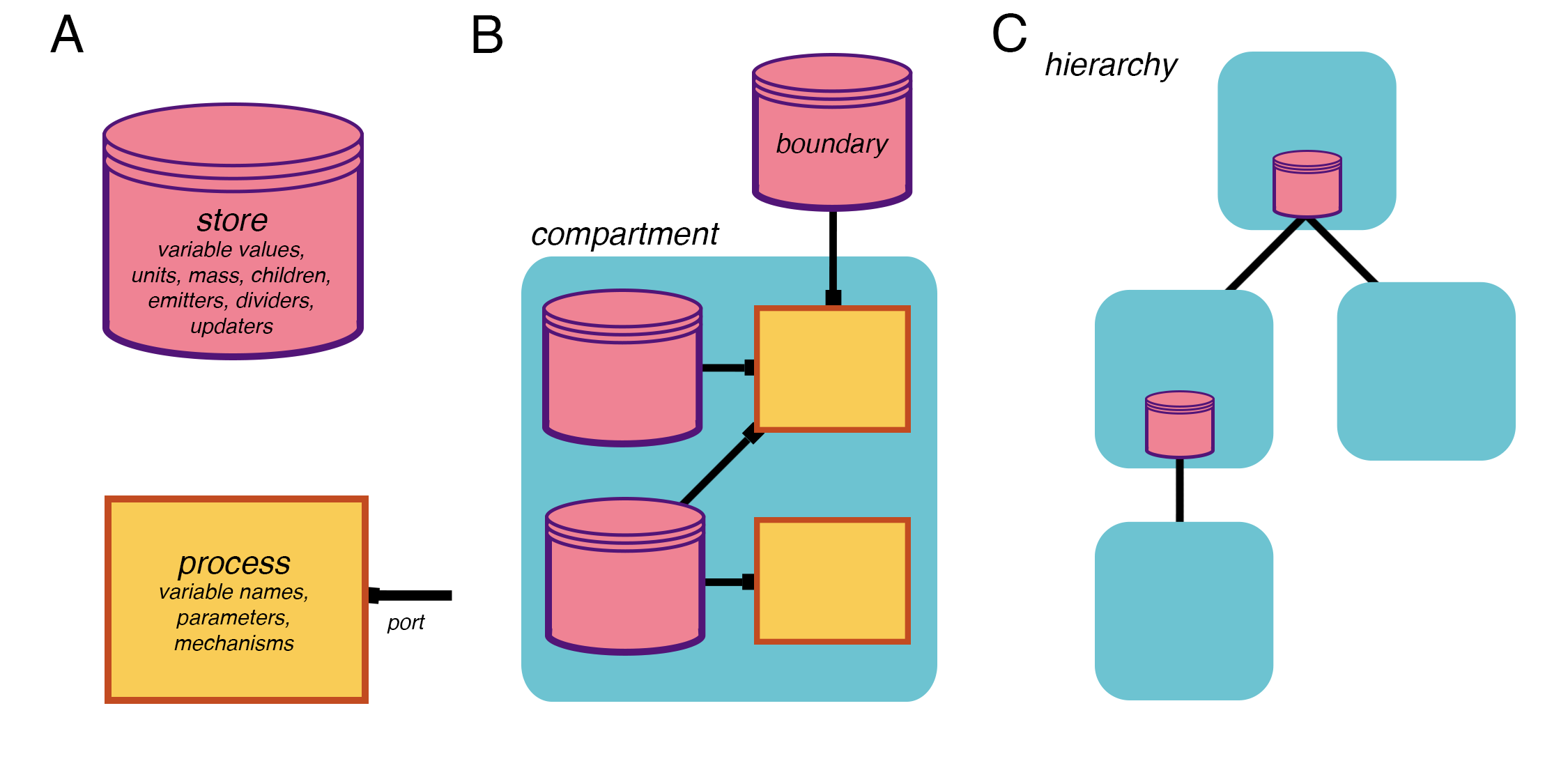
The relationships between stores, processes (panel A), and compartments (panel B) in the tree (panel C).¶
Note that in panel C, only the compartments and boundary stores are shown. The full hierarchy also contains the stores and processes within each compartment.
Hierarchy Structure¶
In the example below, we print out the full hierarchy as a dictionary.
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experimentfrom vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experimentfrom vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import CompartmentExperiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import CompartmentExperiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Experiment
from vivarium.core.process import Compartment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
>>> from vivarium.experiments.glucose_phosphorylation import (
... glucose_phosphorylation_experiment,
... )
>>> from vivarium.core.experiment import Compartment, Experiment
>>> from vivarium.library.pretty import format_dict
>>>
>>>
>>> experiment = glucose_phosphorylation_experiment()
>>> print(format_dict(experiment.state.get_config()))
{
"cell": {
"ADP": {
"_default": 0.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"ATP": {
"_default": 2.0,
"_emit": true,
"_updater": "<function update_accumulate>",
"_value": 2.0
},
"G6P": {
"_default": 0.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.0
},
"GLC": {
"_default": 1.0,
"_emit": true,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 1.0
},
"HK": {
"_default": 0.1,
"_properties": {
"mw": "1.0 gram / mole"
},
"_updater": "<function update_accumulate>",
"_value": 0.1
}
},
"global": {
"initial_mass": {
"_default": "0.0 femtogram",
"_divider": "<function divide_split>",
"_units": "<Unit('femtogram')>",
"_updater": "<function update_set>",
"_value": "0.0 femtogram"
},
"mass": {
"_default": null,
"_emit": true,
"_updater": "<function update_set>",
"_value": "1.826592973891231e-09 femtogram"
}
},
"glucose_phosphorylation": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.glucose_phosphorylation.GlucosePhosphorylation object>"
},
"injector": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.injector.Injector object>"
},
"my_deriver": {
"_default": null,
"_updater": "<function update_set>",
"_value": "<vivarium.processes.tree_mass.TreeMass object>"
}
}
We can represent this hierarchy graphically like this:
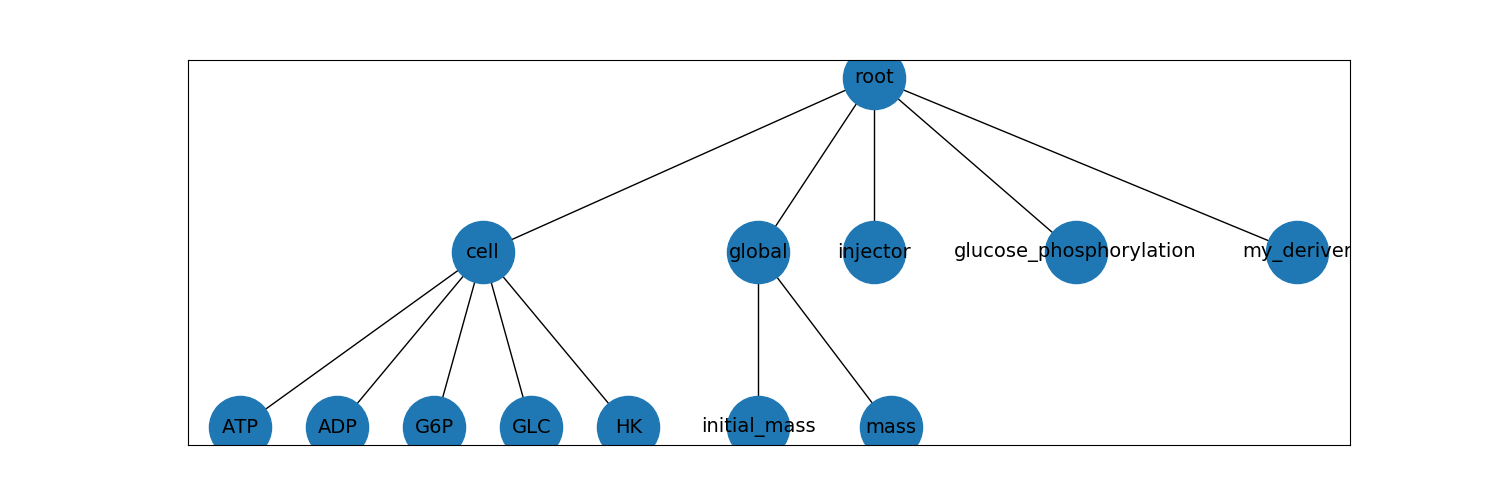
Notice that in the dictionary above, each leaf node in the tree is a key with a value that is a dictionary of schema keys.
Hierarhcy Paths¶
A hierarhcy in Vivarium is like a directory tree on a filesystem. In
line with this analogy, we specify nodes in the hierarchy with paths.
Each path is a tuple of node names (variable names or store names)
relative to some other node. For example, in the topology from the
example above, we used the path ('cell', ) to say that the cell
store maps to the injector’s internal port. This path was
relative to the compartment root (root in our diagram) as is the
case for all topologies. Thus the path is analogous to ./cell in a
directory.
Special Symbols¶
Continuing our analogy between hierarchy paths and file paths, the following symbols have special meanings in hierarchy paths:
..refers to a parent node. One example use for this is a division process that needs to access the parent (environment) compartment to create the daughter cells. In fact, this is what we do in the growth and division compartment:vivarium.compartments.growth_division.*is a wild-card that refers to all the children of a node. For example,(*, )in our topology example above would refer to thecellandglobalstores, as well as theinjector,glucose_phosphorylation, andmy_deriverprocesses. Note that wild-cards don’t make sense in topologies! We just used it here to explain how they work. One example use for wild-cards is in the mass deriver, which uses it to sum masses throughout the hierarchy:vivarium.processes.tree_mass.TreeMass.